Deep learning-based Whole Slide Image (WSI) analysis shows significant potential for automating pathological image diagnosis and intelligent analysis. However, WSIs differ significantly from natural images in size, ranging from 100 million to 10 billion pixels, which prevents the direct application of deep learning models developed for natural images to WSIs. A common approach is to divide WSIs into many non-overlapping small patches for processing, but providing fine-grained annotations for these patches is prohibitively expensive (a WSI can typically produce tens of thousands of patches), rendering patch-based supervised methods impractical. As a result, weakly supervised learning methods based on Multiple Instance Learning (MIL) have become prevalent.
Current challenges in applying MIL to computational pathology include finer-grained classification and localization, multi-scale information fusion, patch and slide-level distribution modeling, elimination of redundancy and ambiguity, and avoiding overfitting, among others.
Our research aims to address these challenges by proposing new, efficient AI algorithms to achieve more accurate, efficient, and intelligent WSI automatic diagnosis and analysis. It is worth mentioning that with the rise of various powerful foundation models, we are actively exploring how to better utilize these models to address various issues in computational pathology.
Weakly/Semi-Supervised Learning Algorithms
-
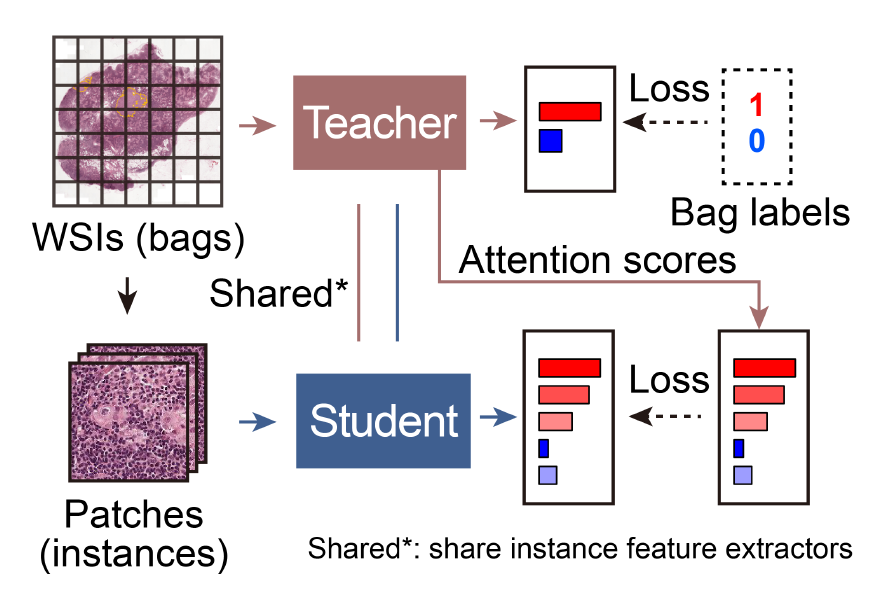
Bi-directional Weakly Supervised Knowledge Distillation for Whole Slide Image Classification
Linhao Qu, Xiaoyuan Luo, Manning Wang, Zhijian Song
Thirty-sixth Conference on Neural Information Processing Systems (
NeurIPS, Spotlight
Presentation in TOP 5%), 2022.
papercode
Abstract▼
Computer-aided pathology diagnosis based on the classification of Whole Slide Image (WSI) plays an important role in clinical practice, and it is often formulated as a weakly-supervised Multiple Instance Learning (MIL) problem. Existing methods solve this problem from either a bag classification or an instance classification perspective. In this paper, we propose an end-to-end weakly supervised knowledge distillation framework (WENO) for WSI classification, which integrates a bag classifier and an instance classifier in a knowledge distillation framework to mutually improve the performance of both classifiers. Specifically, an attention-based bag classifier is used as the teacher network, which is trained with weak bag labels, and an instance classifier is used as the student network, which is trained using the normalized attention scores obtained from the teacher network as soft pseudo labels for the instances in positive bags. An instance feature extractor is shared between the teacher and the student to further enhance the knowledge exchange between them. In addition, we propose a hard positive instance mining strategy based on the output of the student network to force the teacher network to keep mining hard positive instances. WENO is a plug-and-play framework that can be easily applied to any existing attention-based bag classification methods. Extensive experiments on five datasets demonstrate the efficiency of WENO. Code is available at https://github. com/miccaiif/WENO.
-
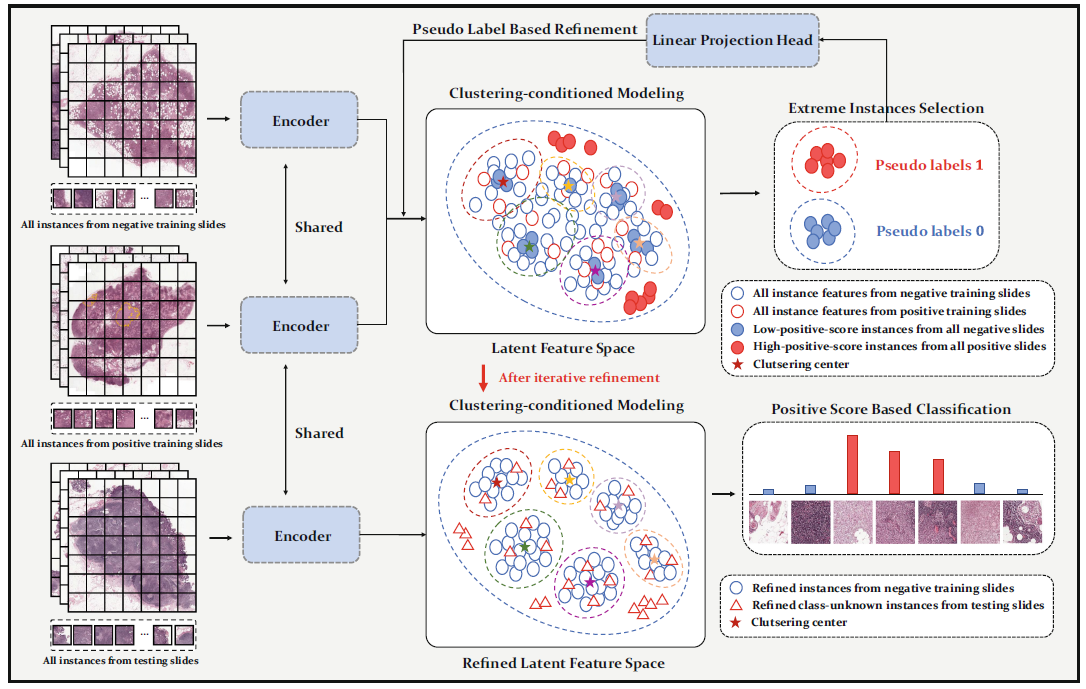
DGMIL: Distribution Guided Multiple Instance Learning for Whole Slide Image Classification
Linhao Qu, xiaoyuan Luo, Shaolei Liu, Manning Wang, Zhijian Song
International Conference on Medical Image Computing and Computer-Assisted Intervention
(
MICCAI), 2022
papercode
Abstract▼
Multiple Instance Learning (MIL) is widely used in analyzing histopathological Whole Slide Images (WSIs). However, existing MIL methods do not explicitly model the data distribution, and instead they only learn a bag-level or instance-level decision boundary discriminatively by training a classifier. In this paper, we propose DGMIL: a feature distribution guided deep MIL framework for WSI classification and positive patch localization. Instead of designing complex discriminative network architectures, we reveal that the inherent feature distribution of histopathological image data can serve as a very effective guide for instance classification. We propose a cluster-conditioned feature distribution modeling method and a pseudo label-based iterative feature space refinement strategy so that in the final feature space the positive and negative instances can be easily separated. Experiments on the CAMELYON16 dataset and the TCGA Lung Cancer dataset show that our method achieves new SOTA for both global classification and positive patch localization tasks.
-
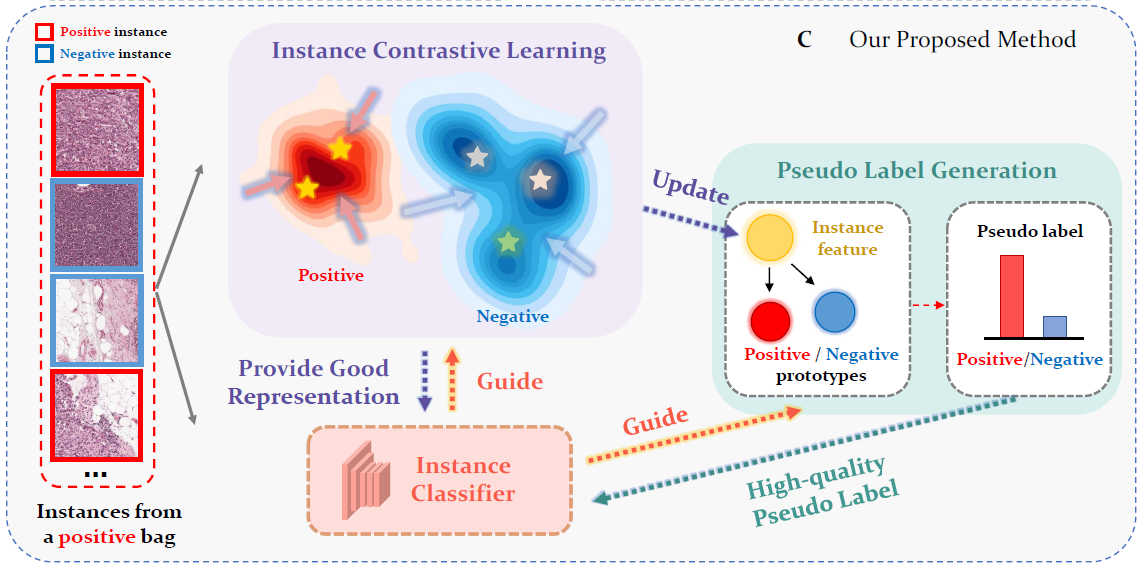
Rethinking Multiple Instance Learning for Whole Slide Image Classification: A Good Instance
Classifier
is All You Need
Linhao Qu, Yingfan Ma, Xiaoyuan Luo, Qinhao Guo, Manning Wang, Zhijian Song
IEEE Transactions on Circuits and Systems for Video Technology (
IF=8.4), 2024.
papercode
Abstract▼
Weakly supervised whole slide image classification is usually formulated as a multiple instance learning (MIL) problem, where each slide is treated as a bag, and the patches cut out of it are treated as instances. Existing methods either train an instance classifier through pseudo-labeling or aggregate instance features into a bag feature through attention mechanisms and then train a bag classifier, where the attention scores can be used for instance-level classification. However, the pseudo instance labels constructed by the former usually contain a lot of noise, and the attention scores constructed by the latter are not accurate enough, both of which affect their performance. In this paper, we propose an instance-level MIL framework based on contrastive learning and prototype learning to effectively accomplish both instance classification and bag classification tasks. To this end, we propose an instance-level weakly supervised contrastive learning algorithm for the first time under the MIL setting to effectively learn instance feature representation. We also propose an accurate pseudo label generation method through prototype learning. We then develop a joint training strategy for weakly supervised contrastive learning, prototype learning, and instance classifier training. Extensive experiments and visualizations on four datasets demonstrate the powerful performance of our method.
-
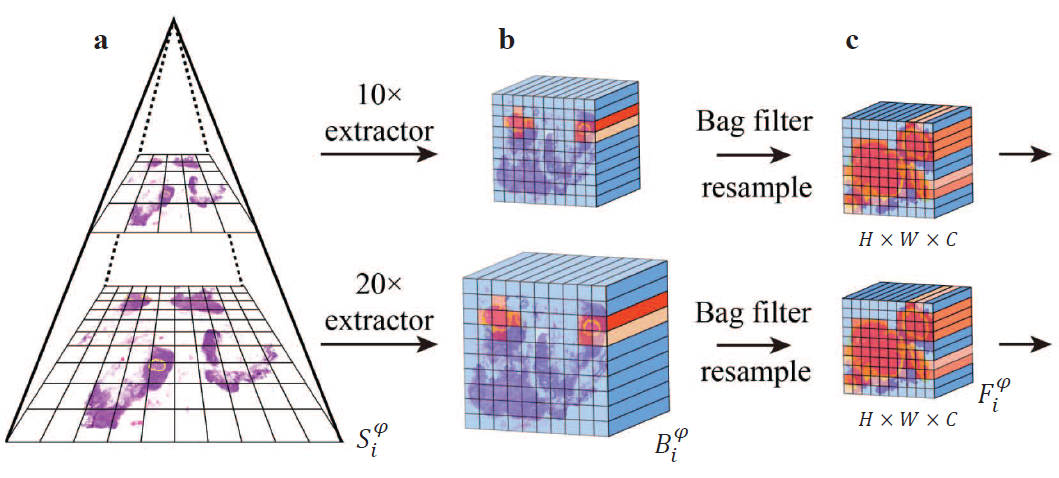
Boosting Whole Slide Image Classification from the Perspectives of Distribution, Correlation and
Magnification
Linhao Qu, Zhiwei Yang, Minghong Duan, Yingfan Ma, Shuo Wang, Manning Wang,
Zhijian
Song
International Conference of Computer Vision (
ICCV), 2023.
papercode
Abstract▼
Bag-based multiple instance learning (MIL) methods have become the mainstream for Whole Slide Image (WSI) classification. However, there are still three important issues that have not been fully addressed: (1) positive bags with a low positive instance ratio are prone to the influence of a large number of negative instances; (2) the correlation between local and global features of pathology images has not been fully modeled; and (3) there is a lack of effective information interaction between different magnifications. In this paper, we propose MILBooster, a powerful dual-scale multi-stage MIL framework to address these issues from the perspectives of distribution, correlation, and magnification. Specifically, to address issue (1), we propose a plug-and-play bag filter that effectively increases the positive instance ratio of positive bags. For issue (2), we propose a novel window-based Transformer architecture called PiceBlock to model the correlation between local and global features of pathology images. For issue (3), we propose a dual-branch architecture to process different magnifications and design an information interaction module called Scale Mixer for efficient information interaction between them. We conducted extensive experiments on four clinical WSI classification tasks using three datasets. MILBooster achieved new state-of-the-art performance on all these tasks.
Foundation Model-Driven Few/Zero-Shot WSI Classification
-
The Rise of AI Language Pathologists: Exploring Two-level Prompt Learning for Few-shot
Weakly-supervised
Whole Slide Image Classification
Linhao Qu, Xiaoyuan Luo, Kexue Fu, Manning Wang, Zhijian Song
Thirty-seventh Conference on Neural Information Processing Systems (
NeurIPS), 2023.
papercode
Abstract▼
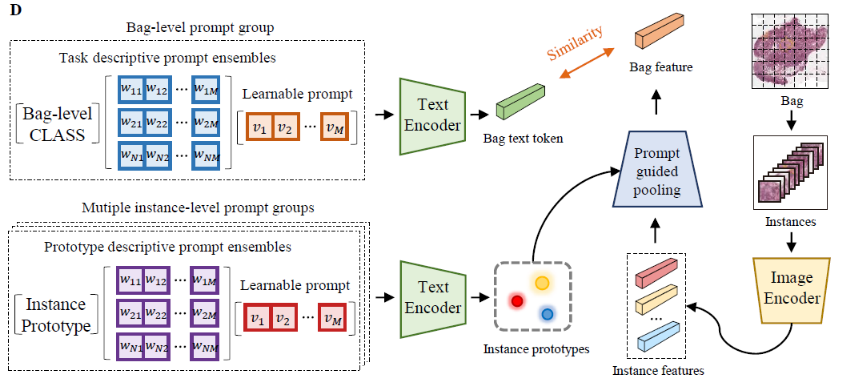
This paper introduces the novel concept of few-shot weakly supervised learning for pathology Whole Slide Image (WSI) classification, denoted as FSWC. A solution is proposed based on prompt learning and the utilization of a large language model, GPT-4. Since a WSI is too large and needs to be divided into patches for processing, WSI classification is commonly approached as a Multiple Instance Learning (MIL) problem. In this context, each WSI is considered a bag, and the obtained patches are treated as instances. The objective of FSWC is to classify both bags and instances with only a limited number of labeled bags. Unlike conventional few-shot learning problems, FSWC poses additional challenges due to its weak bag labels within the MIL framework. Drawing inspiration from the recent achievements of vision-language models (V-L models) in downstream few-shot classification tasks, we propose a two-level prompt learning MIL framework tailored for pathology, incorporating language prior knowledge. Specifically, we leverage CLIP to extract instance features for each patch, and introduce a prompt-guided pooling strategy to aggregate these instance features into a bag feature. Subsequently, we employ a small number of labeled bags to facilitate few-shot prompt learning based on the bag features. Our approach incorporates the utilization of GPT-4 in a question-and-answer mode to obtain language prior knowledge at both the instance and bag levels, which are then integrated into the instance and bag level language prompts. Additionally, a learnable component of the language prompts is trained using the available few-shot labeled data. We conduct extensive experiments on three real WSI datasets encompassing breast cancer, lung cancer, and cervical cancer, demonstrating the notable performance of the proposed method in bag and instance classification. All codes will be made publicly accessible.
-
Pathology-knowledge Enhanced Multi-instance Prompt Learning for Few-shot Whole Slide Image Classification
Linhao Qu, Dingkang Yang, Dan Huang, Qinhao Guo, Rongkui Luo, Shaoting Zhang, and Xiaosong Wang
European Conference on Computer Vision (
ECCV), 2024.
papercode
Abstract▼
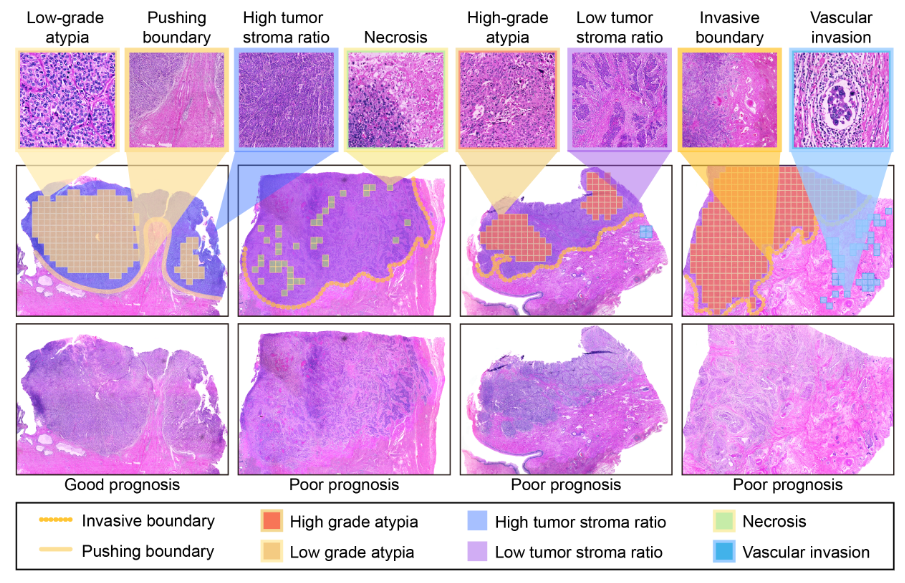
Current multi-instance learning algorithms for pathology image analysis often require a substantial number of Whole Slide Images for effective training but exhibit suboptimal performance in scenarios with limited learning data. In clinical settings, restricted access to pathology slides is inevitable due to patient privacy concerns and the prevalence of rare or emerging diseases. The emergence of the Few-shot Weakly Supervised WSI Classification accommodates the significant challenge of the limited slide data and sparse slide-level labels for diagnosis. Prompt learning based on the pre-trained models (\eg, CLIP) appears to be a promising scheme for this setting; however, current research in this area is limited, and existing algorithms often focus solely on patch-level prompts or confine themselves to language prompts. This paper proposes a multi-instance prompt learning framework enhanced with pathology knowledge, \ie, integrating visual and textual prior knowledge into prompts at both patch and slide levels. The training process employs a combination of static and learnable prompts, effectively guiding the activation of pre-trained models and further facilitating the diagnosis of key pathology patterns. Lightweight Messenger (self-attention) and Summary (attention-pooling) layers are introduced to model relationships between patches and slides within the same patient data. Additionally, alignment-wise contrastive losses ensure the feature-level alignment between visual and textual learnable prompts for both patches and slides. Our method demonstrates superior performance in three challenging clinical tasks, significantly outperforming comparative few-shot methods.
The integration of diverse data modalities—such as images, text, and tabular data—into a unified analytical framework has the potential to significantly enhance medical diagnosis and patient care. Multimodal medical diagnosis leverages the strengths of each data type, providing a comprehensive understanding of patient health and disease progression. While Whole Slide Images (WSIs) offer detailed morphological insights into patient status, cancer progression and responses to treatment are influenced by a multitude of factors. Therefore, incorporating additional modalities, such as radiology images, textual medical reports, and omics data, is crucial for improved outcome prediction.
The primary challenges in this field include efficient multimodal feature extraction, alignment and fusion strategies, multimodal decision-making, and handling missing modalities. These challenges necessitate the development of sophisticated AI algorithms to achieve seamless integration and robust analysis of multimodal data.
Our research focuses on addressing these challenges by proposing novel, efficient AI algorithms. We aim to enhance the accuracy and efficiency of multimodal diagnostic systems through advanced feature extraction and fusion techniques. Additionally, we are actively exploring the application of multimodal foundation models to tackle these challenges, leveraging their vast pre-trained knowledge to improve multimodal data integration and analysis.
Data mining plays a crucial role in extracting valuable insights from multimodal medical data. Our research also emphasizes active learning strategies in medical imaging to optimize data utilization and improve model performance.
Multimodal Medical Diagnosis (Image, Text, Tabular Data)
-
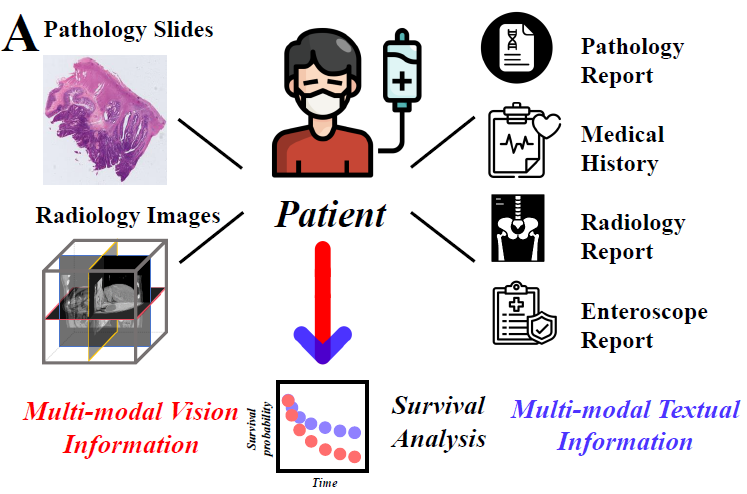
Multi-modal Data Binding for Survival Analysis Modeling with Incomplete Data and Annotations
Linhao Qu,Dan Huang, Shaoting Zhang, and Xiaosong Wang
International Conference on Medical Image Computing and Computer-Assisted Intervention (
MICCAI), 2024.
papercode
Abstract▼
Survival analysis stands as a pivotal process in cancer treatment research, crucial for predicting patient survival rates accurately. Recent advancements in data collection techniques have paved the way for enhancing survival predictions by integrating information from multiple modalities. However, real-world scenarios often present challenges with incomplete data, particularly when dealing with censored survival labels. Prior works have addressed missing modalities but have overlooked incomplete labels, which can introduce bias and limit model efficacy. To bridge this gap, we introduce a novel framework that simultaneously handles incomplete data across modalities and censored survival labels. Our approach employs advanced foundation models to encode individual modalities and align them into a universal representation space for seamless fusion. By generating pseudo labels and incorporating uncertainty, we significantly enhance predictive accuracy. The proposed method demonstrates outstanding prediction accuracy in two survival analysis tasks on both employed datasets. This innovative approach overcomes limitations associated with disparate modalities and improves the feasibility of comprehensive survival analysis using multiple large foundation models.
Active Learning in Medical Imaging
-
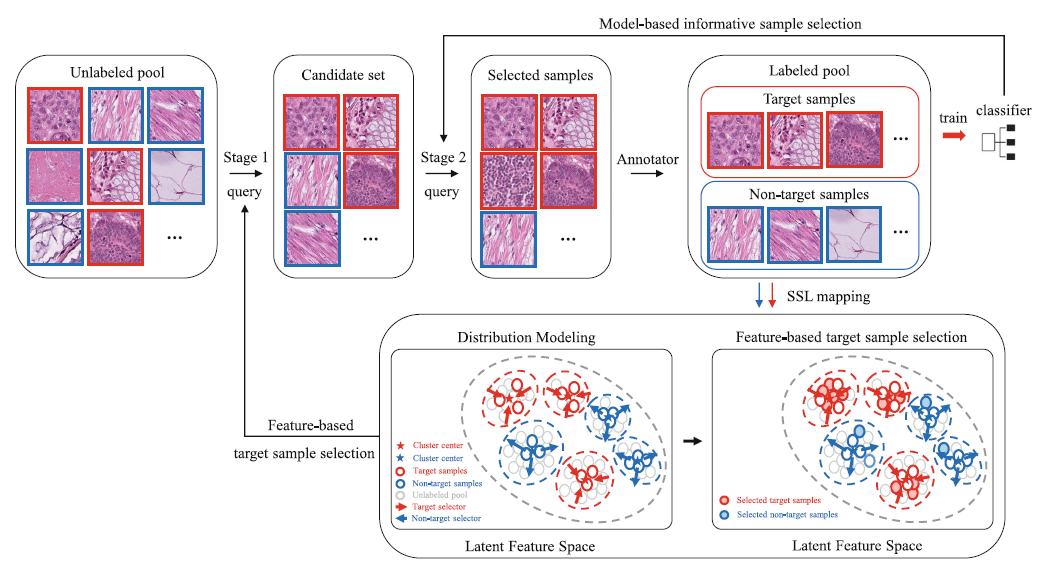
OpenAL: An Efficient Deep Active Learning Framework for Open-Set Histopathology Image
Classification
Linhao Qu, Yingfan Ma, Zhiwei Yang, Manning Wang, Zhijian Song
International Conference on Medical Image Computing and Computer-Assisted Intervention
(
MICCAI), 2023.
paper
code
Abstract▼
Active learning (AL) is an effective approach to select the most informative samples to label so as to reduce the annotation cost. Existing AL methods typically work under the closed-set assumption, i.e., all classes existing in the unlabeled sample pool need to be classified by the target model. However, in some practical clinical tasks, the unlabeled pool may contain not only the target classes that need to be fine-grainedly classified, but also non-target classes that are irrelevant to the clinical tasks. Existing AL methods cannot work well in this scenario because they tend to select a large number of non-target samples. In this paper, we formulate this scenario as an open-set AL problem and propose an efficient framework, OpenAL, to address the challenge of querying samples from an unlabeled pool with both target class and non-target class samples. Experiments on fine-grained classification of pathology images show that OpenAL can significantly improve the query quality of target class samples and achieve higher performance than current state-of-the-art AL methods.
-
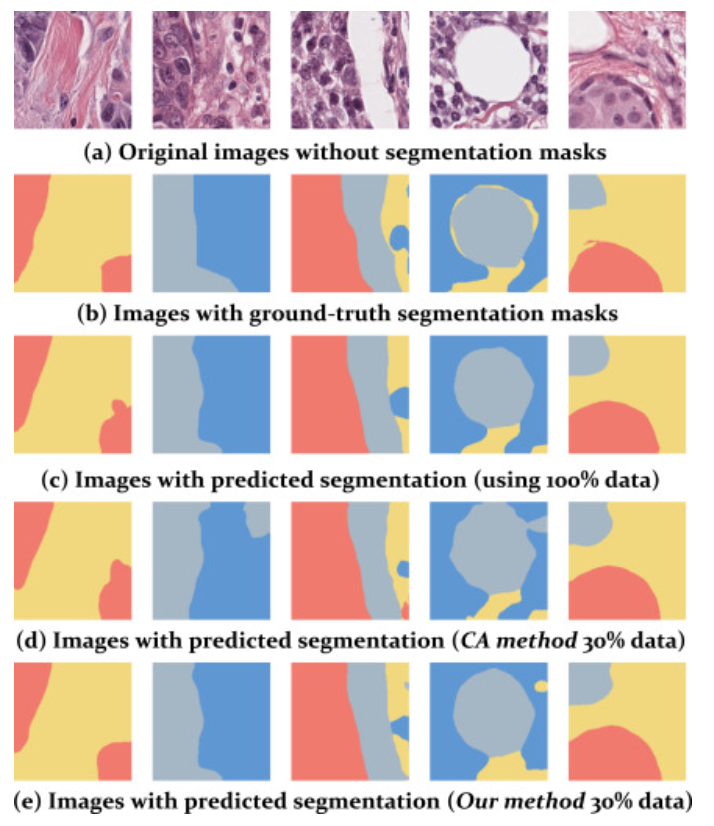
Rethinking deep active learning for medical image segmentation: A diffusion and angle-based framework
Linhao Qu,Qiuye Jin, Kexue Fu, Manning Wang, Zhijian Song
Biomedical Signal Processing and Control (
IF=4.9), 2024.
papercode
Abstract▼
Semantic segmentation based on deep learning typically necessitates a substantial number of finely annotated samples for effective model training. However, acquiring such annotations can be prohibitively expensive and challenging, particularly within the domain of medical image segmentation. Active learning (AL) offers a solution by enabling the selection of the most informative samples for labeling, thereby enhancing annotation efficiency and reducing associated costs. Nevertheless, most existing AL methods operate within an iterative paradigm, which is time-consuming and resource-intensive, thus limiting their practicality in medical applications. Recently, a more viable alternative called the one-shot AL paradigm has emerged, which involves a single interaction with experts and has demonstrated comparable performance to iterative algorithms in image segmentation tasks. In this paper, we propose DifABAL: a novel angle-based one-shot AL framework utilizing diffusion autoencoders for medical image segmentation. Specifically, we leverage diffusion model-based autoencoders to extract features from unlabeled samples and introduce a novel parameter-robust angle-based query strategy for selecting representative samples in the feature space. Extensive experiments conducted on three large publicly available datasets comprising pathology images, chest X-ray images, and dermatology images demonstrate the remarkable performance of our framework.
We are also deeply committed to applying AI algorithms to specific clinical tasks to better address urgent clinical needs. With over two years of experience as research assistant in the pathology department of large comprehensive hospitals, I have progressively applied developed algorithms to areas including gynecology, urology, neuroendocrinology, gastroenterology, head and neck tumors, and rare diseases. We have validated these algorithms with clinical data totaling over 10,000 cases, covering tasks such as tumor diagnosis, prognosis, treatment response prediction, and gene mutation prediction, among others. Concurrently with our research publications, we are also involved in the relevant industrialization process and have applied for multiple related patents.
-
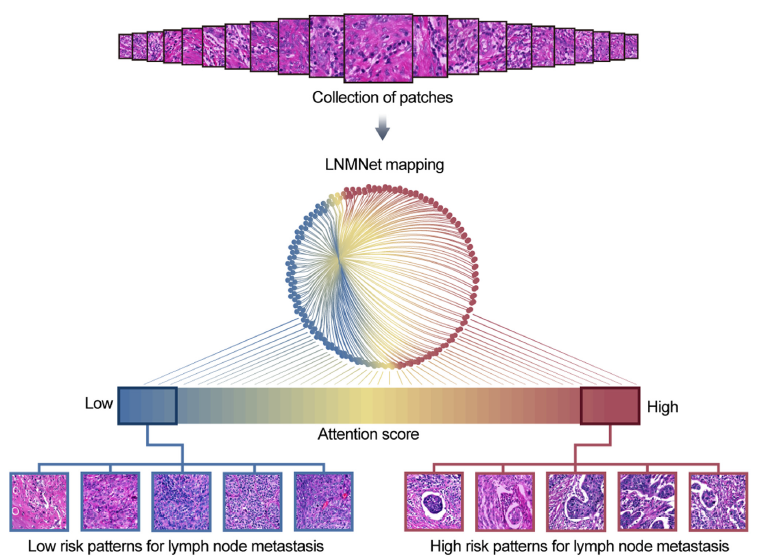
Predicting Lymph Node Metastasis from Primary Cervical Cancer Based on Deep Learning in with
Histopathological Images
Qinhao Guo, Linhao Qu (All technical design and implementation), Jun Zhu, Haiming
Li,
Yong Wu, Simin Wang, Min Yu, Jiangchun Wu, Hao Wen, Xingzhu Ju, Xin Wang, Rui Bi, Yonghong
Shi,
and
Xiaohua Wu
Modern Pathology (
IF=7.5), 2023.
papercode
Abstract▼
We developed a deep learning framework to accurately predict the lymph node status of patients with cervical cancer based on hematoxylin and eosin–stained pathological sections of the primary tumor. In total, 1524 hematoxylin and eosin–stained whole slide images (WSIs) of primary cervical tumors from 564 patients were used in this retrospective, proof-of-concept study. Primary tumor sections (1161 WSIs) were obtained from 405 patients who underwent radical cervical cancer surgery at the Fudan University Shanghai Cancer Center (FUSCC) between 2008 and 2014; 165 and 240 patients were negative and positive for lymph node metastasis, respectively (including 166 with positive pelvic lymph nodes alone and 74 with positive pelvic and para-aortic lymph nodes). We constructed and trained a multi-instance deep convolutional neural network based on a multiscale attention mechanism, in which an internal independent test set (100 patients, 228 WSIs) from the FUSCC cohort and an external independent test set (159 patients, 363 WSIs) from the Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma cohort of the Cancer Genome Atlas program database were used to evaluate the predictive performance of the network. In predicting the occurrence of lymph node metastasis, our network achieved areas under the receiver operating characteristic curve of 0.87 in the cross-validation set, 0.84 in the internal independent test set of the FUSCC cohort, and 0.75 in the external test set of the Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma cohort of the Cancer Genome Atlas program. For patients with positive pelvic lymph node metastases, we retrained the network to predict whether they also had para-aortic lymph node metastases. Our network achieved areas under the receiver operating characteristic curve of 0.91 in the cross-validation set and 0.88 in the test set of the FUSCC cohort. Deep learning analysis based on pathological images of primary foci is very likely to provide new ideas for preoperatively assessing cervical cancer lymph node status; its true value must be validated with cervical biopsy specimens and large multicenter datasets.
-
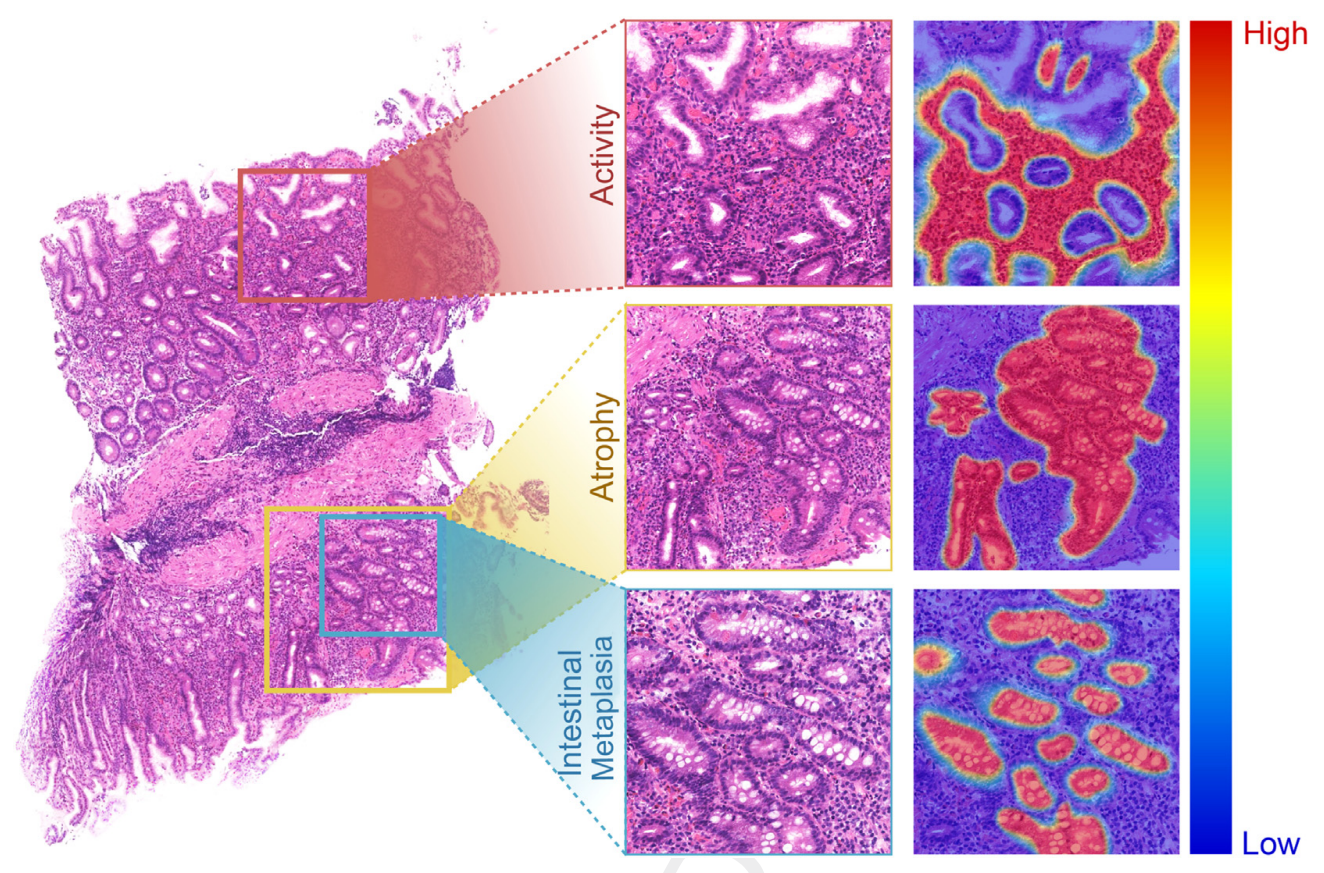
Advancing Automatic Gastritis Diagnosis: An Interpretable Multilabel Deep Learning Framework for
the
Simultaneous Assessment of Multiple Indicators
Mengke Ma, Xixi Zeng, Linhao Qu (All technical design and implementation), Xia
Sheng, Hongzheng Ren, Weixiang Chen, Bin Li, Qinghua You, Li Xiao, Yi Wang, Mei Dai, Boqiang
Zhang,
Changqing Lu, Weiqi Sheng, and Dan Huang
The American Journal of Pathology (
IF=6.0), 2024.
papercode
Abstract▼
The evaluation of morphologic features, such as inflammation, gastric atrophy, and intestinal metaplasia, is crucial for diagnosing gastritis. However, artificial intelligence analysis for nontumor diseases like gastritis is limited. Previous deep learning models have omitted important morphologic indicators and cannot simultaneously diagnose gastritis indicators or provide interpretable labels. To address this, an attention-based multi-instance multilabel learning network (AMMNet) was developed to simultaneously achieve the multilabel diagnosis of activity, atrophy, and intestinal metaplasia with only slide-level weak labels. To evaluate AMMNet's real-world performance, a diagnostic test was designed to observe improvements in junior pathologists' diagnostic accuracy and efficiency with and without AMMNet assistance. In this study of 1096 patients from seven independent medical centers, AMMNet performed well in assessing activity [area under the curve (AUC), 0.93], atrophy (AUC, 0.97), and intestinal metaplasia (AUC, 0.93). The false-negative rates of these indicators were only 0.04, 0.08, and 0.18, respectively, and junior pathologists had lower false-negative rates with model assistance (0.15 versus 0.10). Furthermore, AMMNet reduced the time required per whole slide image from 5.46 to 2.85 minutes, enhancing diagnostic efficiency. In block-level clustering analysis, AMMNet effectively visualized task-related patches within whole slide images, improving interpretability. These findings highlight AMMNet's effectiveness in accurately evaluating gastritis morphologic indicators on multicenter data sets. Using multi-instance multilabel learning strategies to support routine diagnostic pathology deserves further evaluation.











